Automated detection of chromosome structure and determination of cell ploidy remains an elusive goal owing to the diverse variations in optical chromosome shape. While the number of chromosomes and chromosome identities remain conserved across healthy individuals, imaging variations in chromosome karyotypes necessitate the experience of a pathologist or geneticist to validate chromosome count. These variations arise from not only from debris arising from preparation of chromosomal fixation on glass slides, but also from varying degrees of overlap and separation between individual chromosomes. Counting of chromosomes is further exacerbated by instances of chromosomal instability, where cells possess an abnormal number of chromosomes resulting from nondisjunction during meiosis. Detection of aneuploidy is difficult as variance in aneuploidy ranges from loss or gain of any number of chromosomes, emphasizing the need for both an accurate and precise model. An automated method for counting overlapping chromosome clusters using computer vision with machine learning will be useful for any labs looking to study aneuploidy in a high throughput manner. Below we present a chromosome counting model that is comprised of three stages: (1) chromosome isolation from background artifacts and debris, (2) identification and inference of individual chromosome counts in clumps, followed by (3) unsupervised clustering of chromosome groups on a per cell basis.
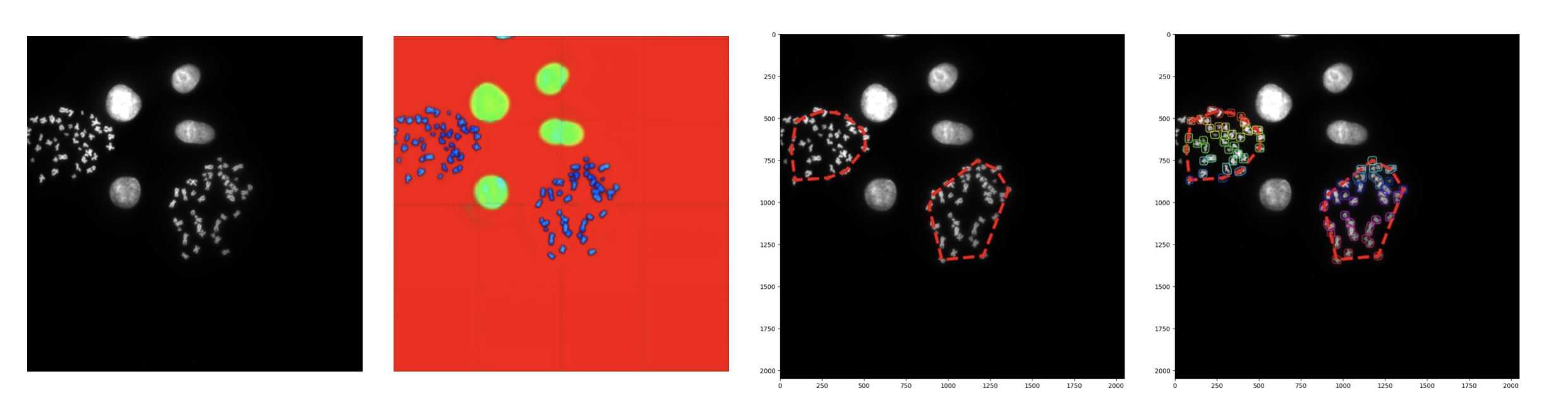
When integrated into the standard sample preparation and microscopy workflow, this model will improve data processing turnaround time from when the sample is collected to when it is quantitatively analyzed. Existing platforms such as ThermoFisher’s Eureka Analysis, can identify areas of interest on microscope slide, but are unable to provide immediate quantitative feedback. This model when developed can act as a module such that when integrated into the vision module of the microscope directly can inform the scientist the characteristics of the sample.
This project was developed using Tensorflow, with deployment executed via Docker and Flask.